Visualization
Visualization is an optional, but desirable aspect of tasks. Often,
visualization is an optional step of a task but can also be included
as part of other steps such as init or analysis.
While developers can write their own visualization scripts associated with
individual tasks, the following shared visualization routines are
provided in polaris.viz:
common matplotlib style
The function polaris.viz.use_mplstyle() loads a common
matplotlib style sheet
that can be used to make font sizes and other plotting options more consistent
across Polaris. The plotting functions described below make use of this common
style. Custom plotting should call polaris.viz.use_mplstyle()
before creating a matplotlib figure.
horizontal fields from planar meshes
polaris.viz.plot_horiz_field() produces a patches-style
visualization of x-y fields across a single vertical level at a single time
step. The image file (png) is saved to the directory from which
polaris.viz.plot_horiz_field() is called. The function
automatically detects whether the field specified by its variable name is
a cell-centered variable or an edge-variable and generates the patches, the
polygons characterized by the field values, accordingly.
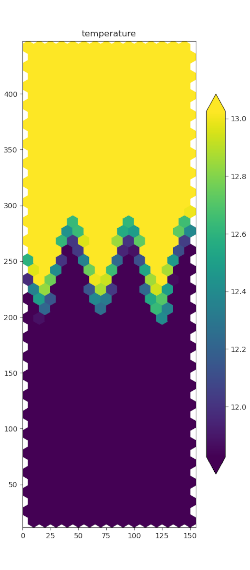

An example function call that uses the default vertical level (top) is:
cell_mask = ds_init.maxLevelCell >= 1
plot_horiz_field(config, ds, ds_mesh, 'normalVelocity',
'final_normalVelocity.png',
t_index=t_index,
vmin=-max_velocity, vmax=max_velocity,
cmap='cmo.balance', show_patch_edges=True,
cell_mask=cell_mask)
The cell_mask argument can be any field indicating which horizontal cells
are valid and which are not. A typical value for ocean plots is as shown
above: whether there are any active cells in the water column.
For increased efficiency, you can store the patches and patch_mask from
one call to plot_horiz_field() and reuse them in subsequent calls. The
patches and patch_mask are specific to the dimension (nCell or nEdges)
of the field to plot and the cell_mask. So separate patches and
patch_mask variables should be stored for as needed:
cell_mask = ds_init.maxLevelCell >= 1
cell_patches, cell_patch_mask = plot_horiz_field(
ds=ds, ds_mesh=ds_mesh, field_name='ssh', out_file_name='plots/ssh.png',
vmin=-720, vmax=0, figsize=figsize, cell_mask=cell_mask)
plot_horiz_field(ds=ds, ds_mesh=ds_mesh, field_name='bottomDepth',
out_file_name='plots/bottomDepth.png', vmin=0, vmax=720,
figsize=figsize, patches=cell_patches,
patch_mask=cell_patch_mask)
edge_patches, edge_patch_mask = plot_horiz_field(
ds=ds, ds_mesh=ds_mesh, field_name='normalVelocity',
out_file_name='plots/normalVelocity.png', t_index=t_index,
vmin=-0.1, vmax=0.1, cmap='cmo.balance', cell_mask=cell_mask)
...
global lat/lon plots
plotting from spherical MPAS meshes
You can use polaris.viz.plot_global_mpas_field() to plot a field on
a spherical MPAS mesh.

Typical usage might be:
import cmocean # noqa: F401
import xarray as xr
from polaris import Step
from polaris.viz import plot_global_mpas_field
class Viz(Step):
def run(self):
ds = xr.open_dataset('initial_state.nc')
da = ds['tracer1'].isel(Time=0, nVertLevels=0)
plot_global_mpas_field(
mesh_filename='mesh.nc', da=da,
out_filename='init.png', config=self.config,
colormap_section='cosine_bell_viz',
title='Tracer at init', plot_land=False,
central_longitude=180.)
The plot_land parameter to polaris.viz.plot_global_mpas_field() is
used to enable or disable continents overlain on top of the data.
The central_longitude defaults to 0.0 and can be set to another value
(typically 180 degrees) for visualizing quantities that would otherwise be
divided across the antimeridian.
The colormap_section of the config file must contain config options for
specifying the colormap:
# options for visualization for the cosine bell convergence test case
[cosine_bell_viz]
# colormap options
# colormap
colormap_name = viridis
# the type of norm used in the colormap
norm_type = linear
# colorbar limits
colorbar_limits = 0.0, 1.0
colormap_name can be any available matplotlib colormap. For ocean test
cases, we recommend importing cmocean so
the standard ocean colormaps are available.
The norm_type is one of linear (a linear colormap) or log (a logarithmic
colormap).
The colorbar_limits are the lower and upper bound of the colorbar range.
plotting from lat/lon grids
You can use polaris.viz.plot_global_lat_lon_field() to plot a field
on a regular lon-lat grid, perhaps after remapping from an MPAS mesh using
polaris.remap.MappingFileStep.

The plot_land parameter to polaris.viz.plot_global_lat_lon_field()
is used to enable or disable continents overlain on top of the data:

Typical usage might be:
import cmocean # noqa: F401
import xarray as xr
from polaris import Step
from polaris.viz import plot_global_lat_lon_field
class Viz(Step):
def run(self):
ds = xr.open_dataset('initial_state.nc')
ds = ds[['tracer1']].isel(Time=0, nVertLevels=0)
plot_global_lat_lon_field(
ds.lon.values, ds.lat.values, ds.tracer1.values,
out_filename='init.png', config=self.config,
colormap_section='cosine_bell_viz',
title='Tracer at init', plot_land=False)
The colormap_section of the config file must contain config options for
specifying the colormap:
# options for visualization for the cosine bell convergence task
[cosine_bell_viz]
# colormap options
# colormap
colormap_name = viridis
# the type of norm used in the colormap
norm_type = linear
# A dictionary with keywords for the norm
norm_args = {'vmin': 0., 'vmax': 1.}
# We could provide colorbar tick marks but we'll leave the defaults
# colorbar_ticks = np.linspace(0., 1., 9)
colormap_name can be any available matplotlib colormap. For ocean test
cases, we recommend importing cmocean so
the standard ocean colormaps are available.
The norm_type is one of linear (a linear colormap), symlog (a
symmetric log
colormap with a central linear region), or log (a logarithmic colormap).
The norm_args depend on the norm_typ and are the arguments to
matplotlib.colors.Normalize, matplotlib.colors.SymLogNorm,
and matplotlib.colors.LogNorm, respectively.
The config option colorbar_ticks (if it is defined) specifies tick locations
along the colorbar. If it is not specified, they are determined automatically.